Guest post by Philipp Schlegel, Cambridge Connectomics/Jefferis Lab
Dear FlyWire community,
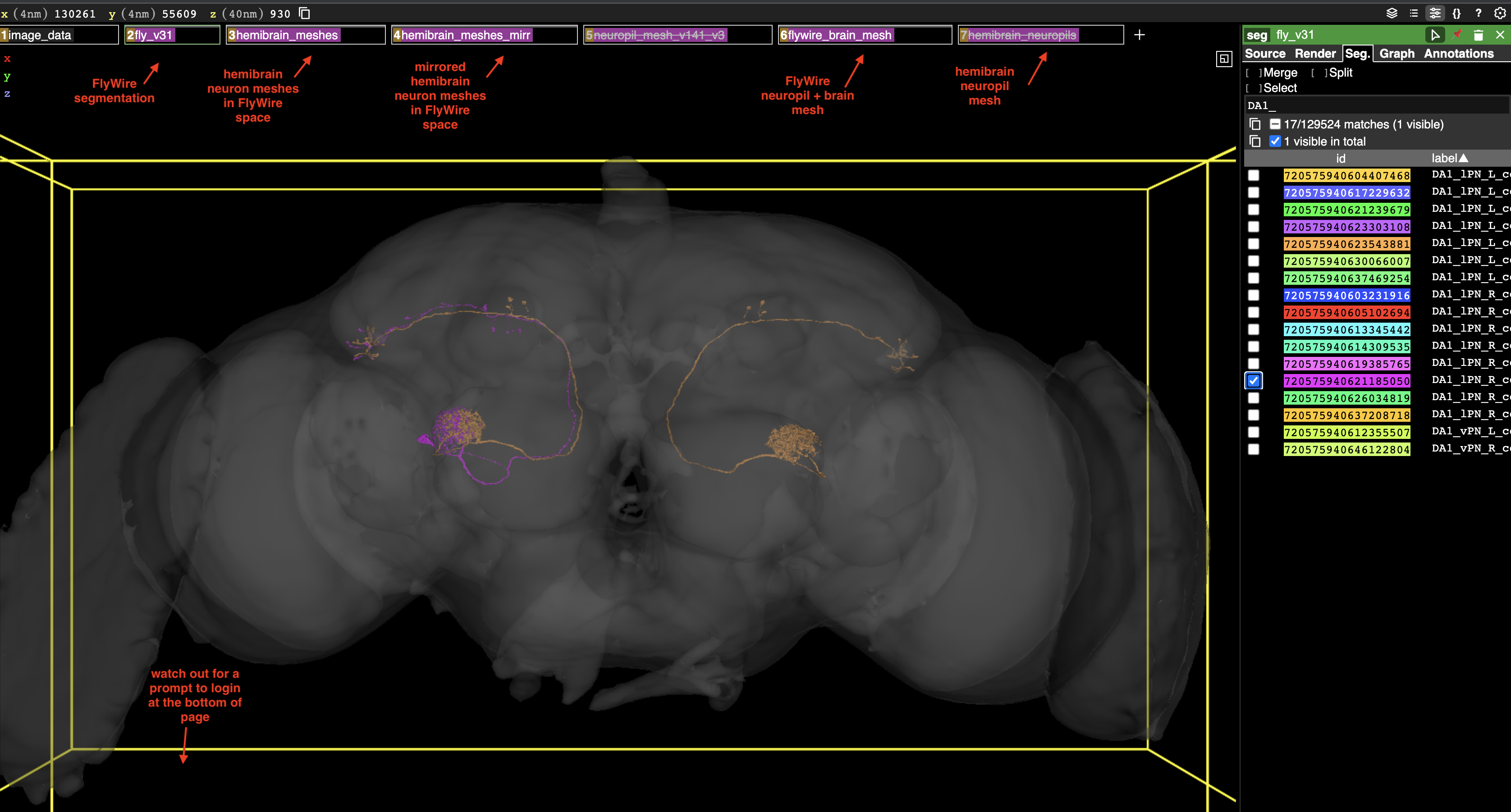
As promised during the town hall meeting on March 30, 2023, here is a neuroglancer link with a couple layers you might find useful:
- The FlyWire segmentation layer contains searchable labels generated from the hierarchical annotations – this should ~ match what you see in Codex. There are two technical limitations you need to be aware of: (a) the mapping between root IDs and the labels is updated every 30mins which means edits are tracked but not instantly; (b) neuroglancer caches the labels which means you need to manually reload the website to refresh them. Since most neurons are pretty stable at this point I don’t anticipate huge issues with this. Once there is a static segmentation for the
630release version this will become a non-issue. Also note that in this base neuroglancer, the FlyWire login prompt will appear at the bottom of the page! - Two layers for hemibrain neuron meshes in FlyWire space: one mirrored and one un-mirrored. These also have searchable labels based on the
instancefield from neuPrint. - A layer for the hemibrain neuropil mesh in FlyWire space.
- A layer with a new FlyWire brain and neuropil mesh (see screenshot). These were generated from the segmentation tissue mask and the synapse cloud, respectively, and as such are a better fit than the older meshes.
The annotation labels won’t work with the FlyWire neuroglancer (yet) but you should be able to drag & drop the other layers into FlyWire neuroglancer.
We kindly ask you not to share these data beyond the FlyWire community for now.
Hope you’ll find this useful and please reach out to Philipp Schlegel in FlyWire Slack if you run into issues!