Here are some proofreading tips specific to the BANC (Brain and Nerve Cord) dataset in FlyWire.
Using Mitochondria For Continuations
For this particular dataset, you may often find that the dark mitochondria in a branch cause the AI to grow confused and terminate the branch instead of finding the extension. However, although most mitochondria don’t need to be added to what you’re proofreading, you can use them to your advantage.
When dealing with a main branch that’s cut off from where it should obviously continue, try selecting a dark mitochondrial oval in the 2D EM view, and then look ahead for what seems wrapped around it. This is very likely the continuation of your original branch.
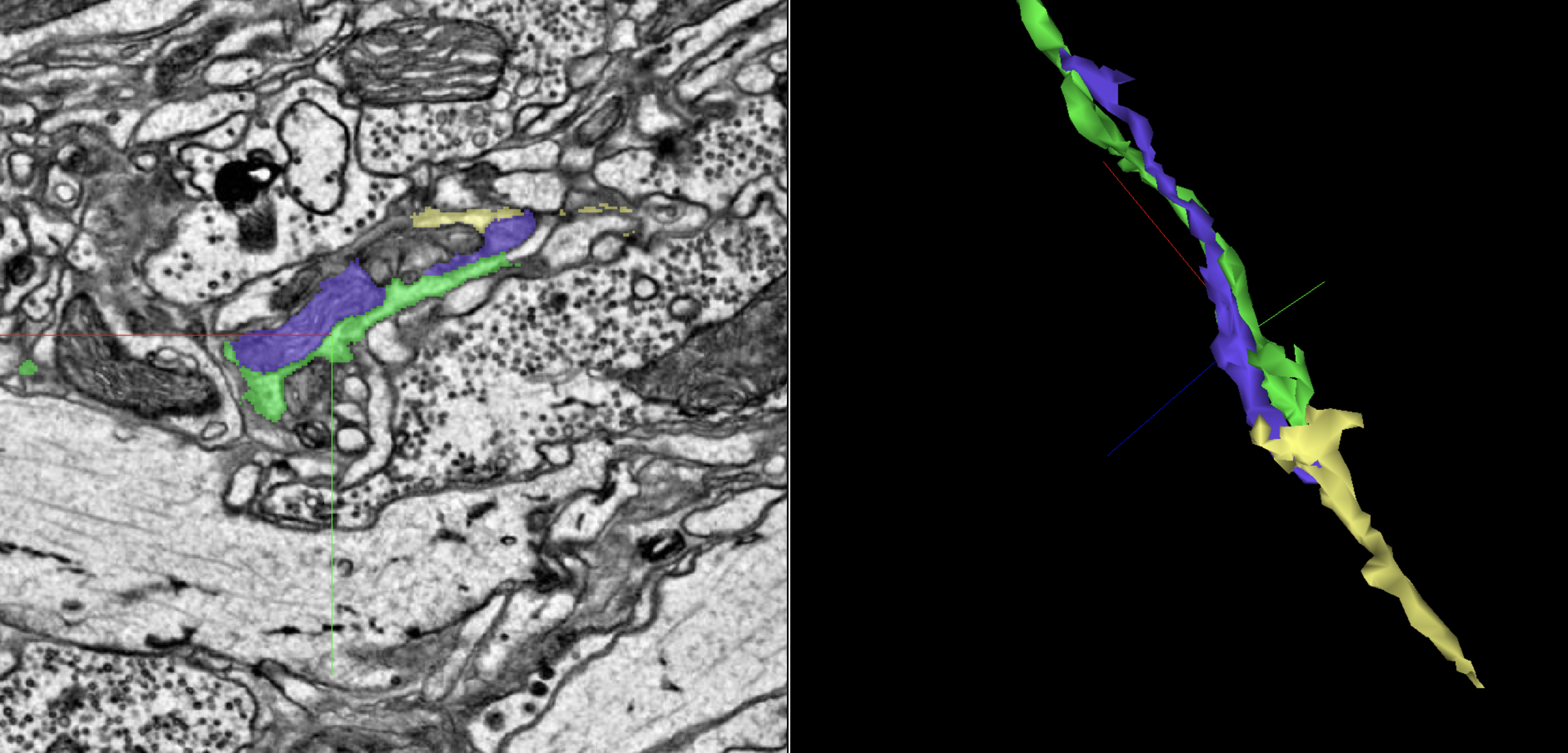
In the above image, the green segment appears to cut off, but the purple mitochondria shows the way to the gold extension.
It’s up to you whether to include the mitochondria in your final revision.
X-Shaped, Perpendicular Mergers
These are a classic merger type that will be familiar for FlyWire users. If your chosen branch is crossed by another branch at a 90º angle, or close enough, this perpendicular formation suggests a merger exists. It’s usually very simple to place multicut points on each branch and separate them.
Sometimes these mergers will be less apparent if the invading branch also cuts off before it can form a complete X, instead forming something more like a T. When examining a T intersection between branches, if they’re all on exactly the same plane then they’re probably correct; but if one seems to be tacked on from a slightly different plane, take a closer look since it may still be a perpendicular merger.
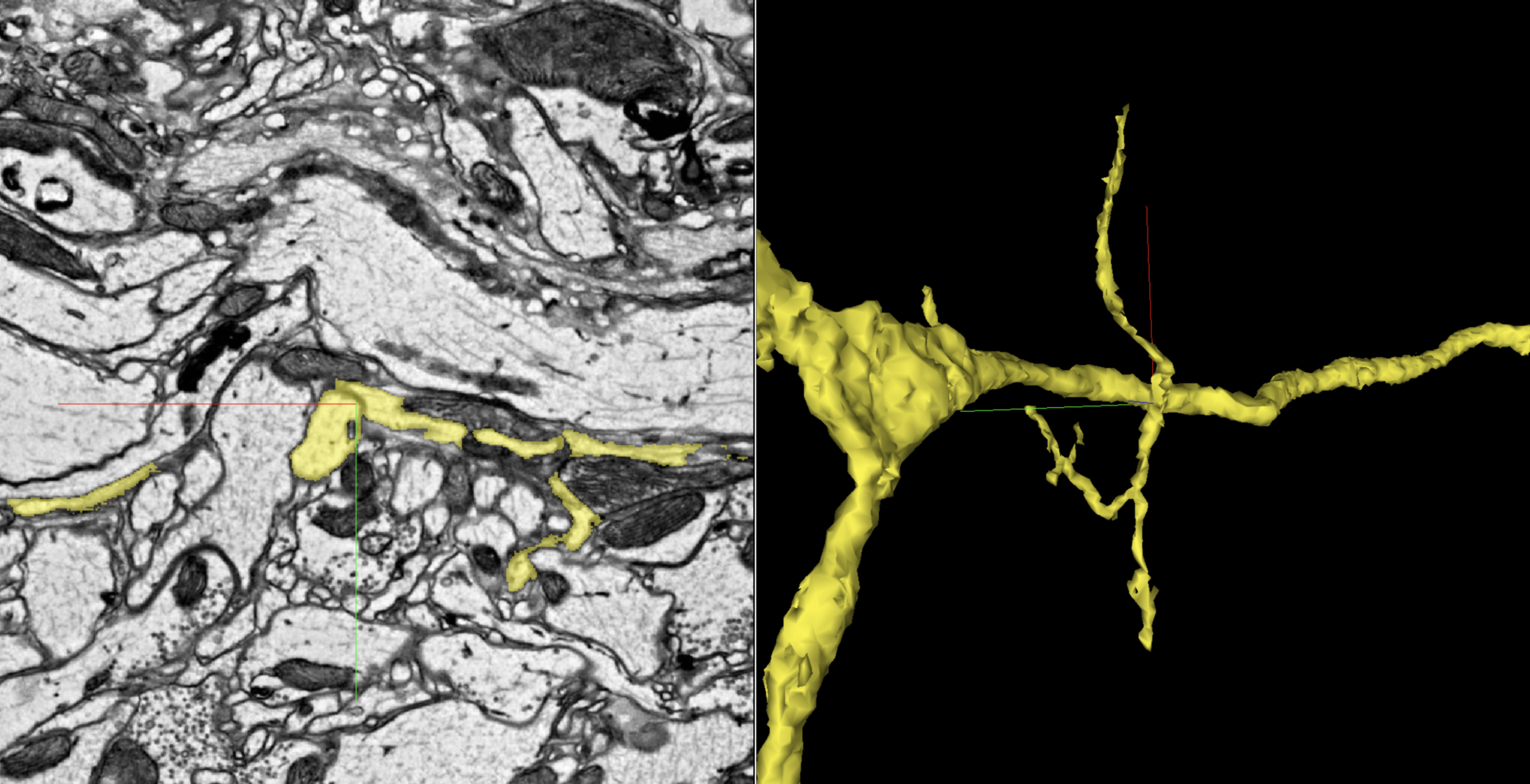
As you can see in the below image, this is definitely some erroneous criss-crossing. On the left, the crosshairs center on where a fairly solid boundary was ignored.
H-Shaped, Parallel Mergers
Another familiar merger type, where two branches run directly in parallel with each other and touch for a good portion of that distance. However, while it may be very obvious to spot, you may need several tries with multicut to split the two branches where they’ve actually merged. Make sure you’ve placed your points on one of those direct contact areas.
As with perpendicular mergers, parallel mergers can sometimes hide in plain sight with a very small area where the two branches meet before both branches cut off in opposite directions. This may suggest a normal continuation when zoomed out in 3D. Try to keep your 3D reasonably zoomed in when proofreading.
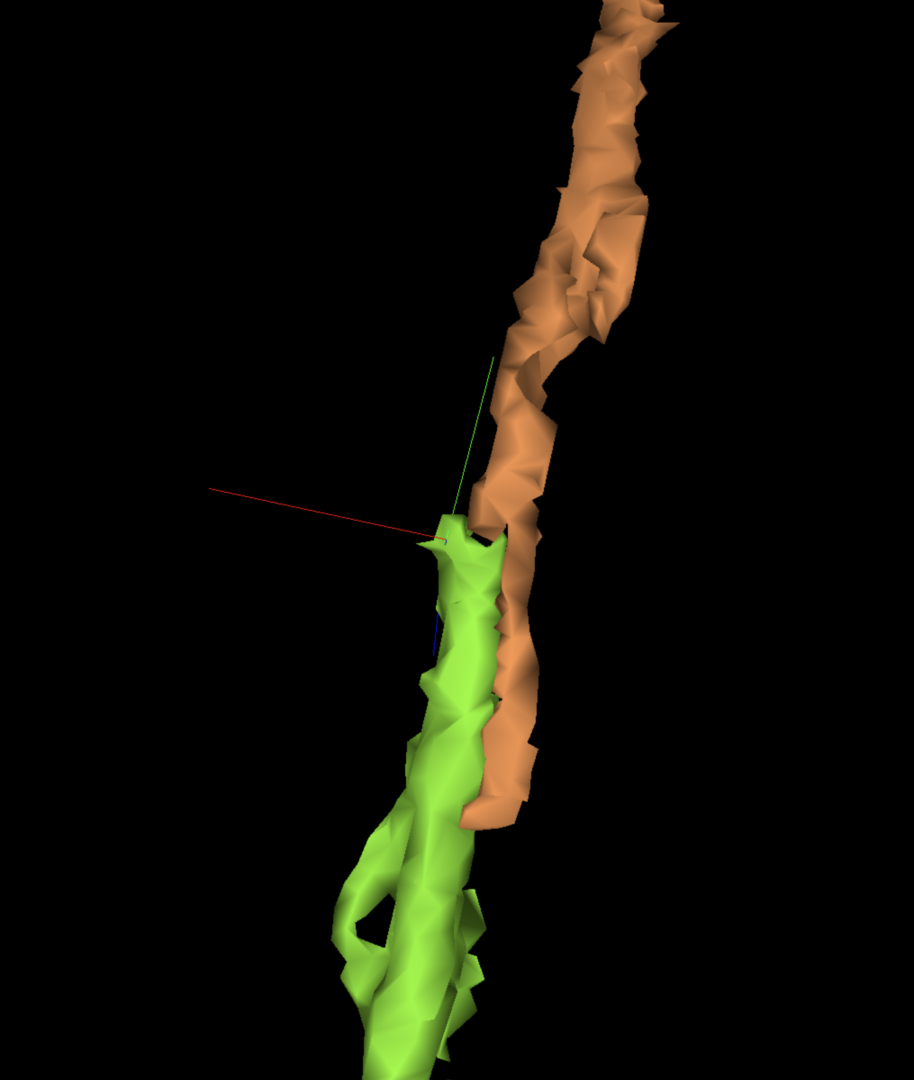
For more on parallel merger tendencies, see “Path Swaps” below.
Misalignments & Slide Pinches
The BANC dataset has plenty of examples of what may feel like a familiar challenge for interpreting any EM slides. At certain points the slides have been scanned in such a way that one slide slipped out of alignment with the ones before it. This misalignment appears as a “jump” where it can be hard, especially for the AI, to keep track of where a particular segment should flow into another.
There are also instances of slides being pinched or crumpled and causing similar interruptions as the AI cannot navigate past them.
Just like on FlyWire, you can use the annotation tool to help yourself keep track of a segment before and after any misalignment or pinch:
- Find a unique formation that stands out in the 2D EM view, and make sure you can spot it in the EM both before and after the slide jumps or pinches.
- Add annotations to this formation both before and after the jump/pinch.
- By using these annotations as frames of reference, you can more easily find where your chosen neuron continues in relation to its surroundings.
For some of these situations, the above procedure may not always be enough, in which case the following video for the original FlyWire dataset can still be of help:
Lastly, take note that in this dataset there are a few areas in which the slides were particularly folded or pinched, and these show as thick black bars cutting across most of the EM. You will frequently need to continue a branch across these black bars, for which you may want to combine misalignment annotation tricks and following mitochondria.
Gaps in Dendritic Arbors
In BANC, the dendritic arbor of most cells will be complete enough with just the AI that you may not need to add much to this part of the cell as you proofread. Your lab may also not be interested in viewing complete dendritic arbors.
However, it’s still possible for the AI to miss larger dendrites, which you may find by reviewing the arbor’s overall morphology and checking for voids in otherwise consistent fractal branches. Keep an eye out for these missing formations, and consult with your team about how thoroughly you need to fill things in.
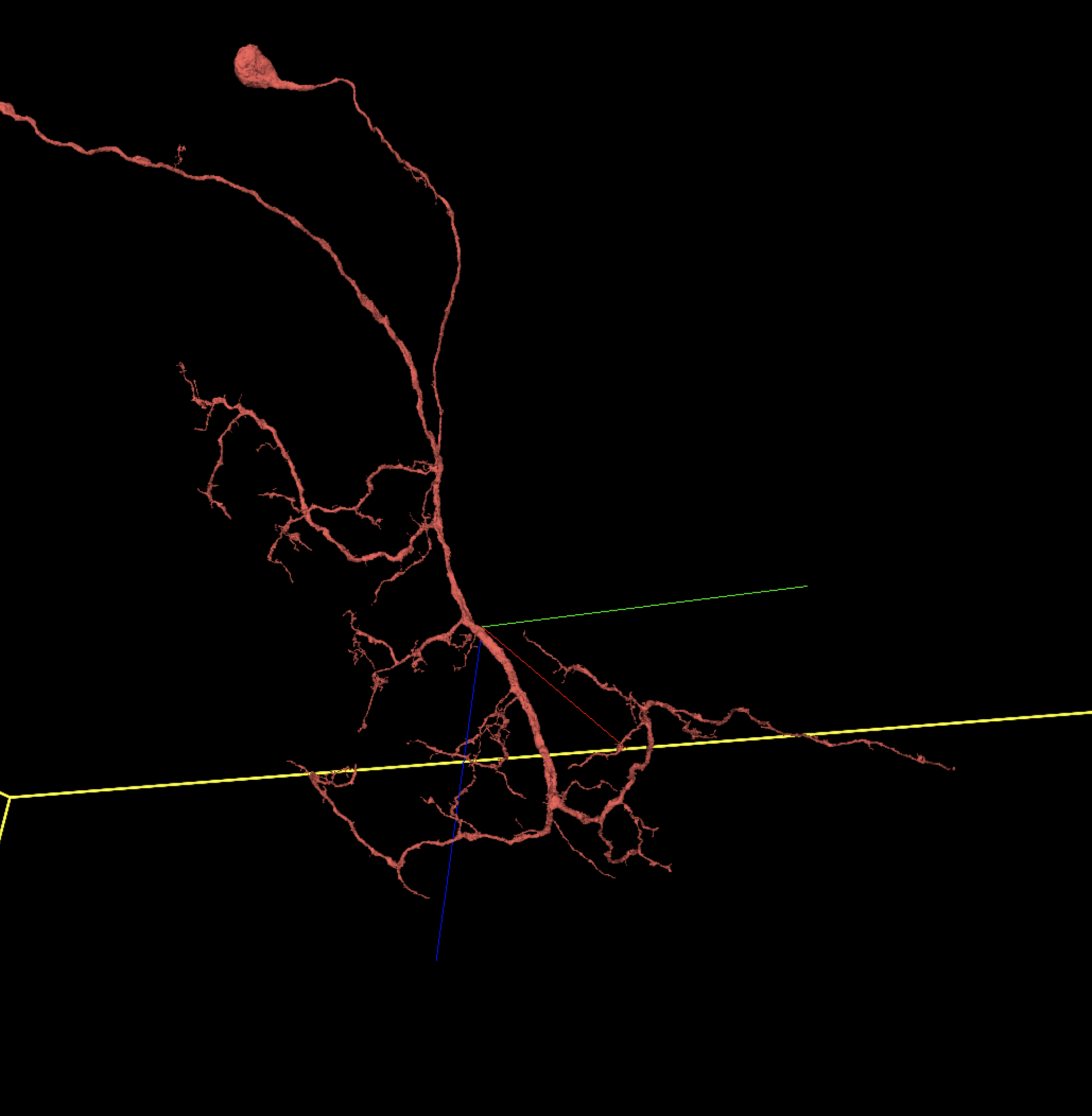
Some dendrites will be much denser and branch more plentifully than the one above, but many may look as sparse as this — however, they should still be examined for relative evenness.
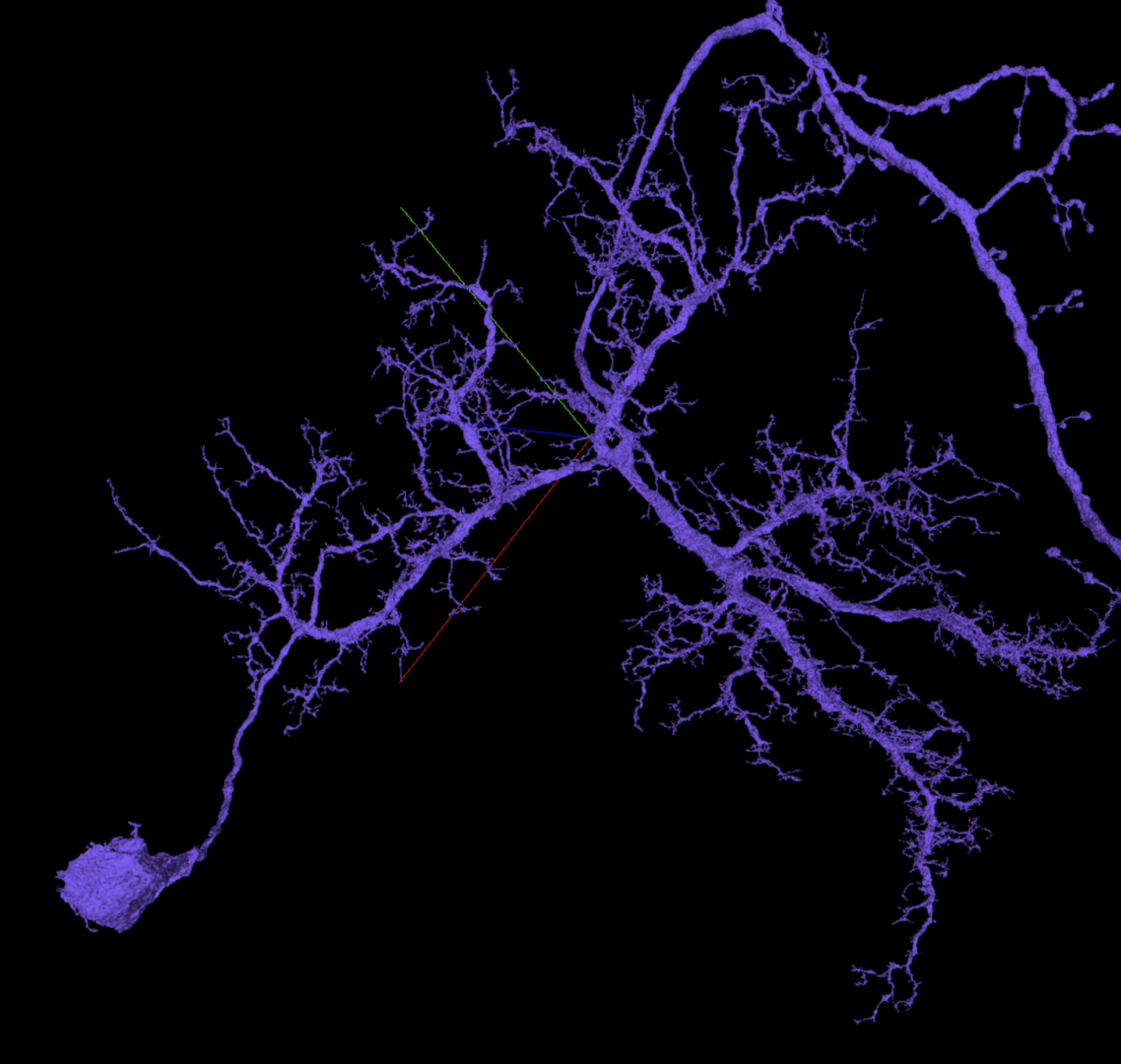
Here is a different dendritic arbor that’s much more complicated. Note that branches in the upper right corner are axonal.
Axonal Boutons & Other Thin Extensions
Many branch continuations can be extremely narrow due to neurons being packed in tightly within one area, or due to the cell anatomy becoming very delicate. In BANC you’ll especially find this happening with boutons joined to an axonal terminal.
To find missing boutons for an axon, first look in the 3D for a thin, raised bump on a branch that suggests a twig shape emerging. In the 2D, scan for small, nearly self-contained blobs floating near that bump; selecting one of these will often show you a bouton that just barely connects to the bump you started at. If it isn’t a full connection but the pathway looks clear enough, you should be all set.
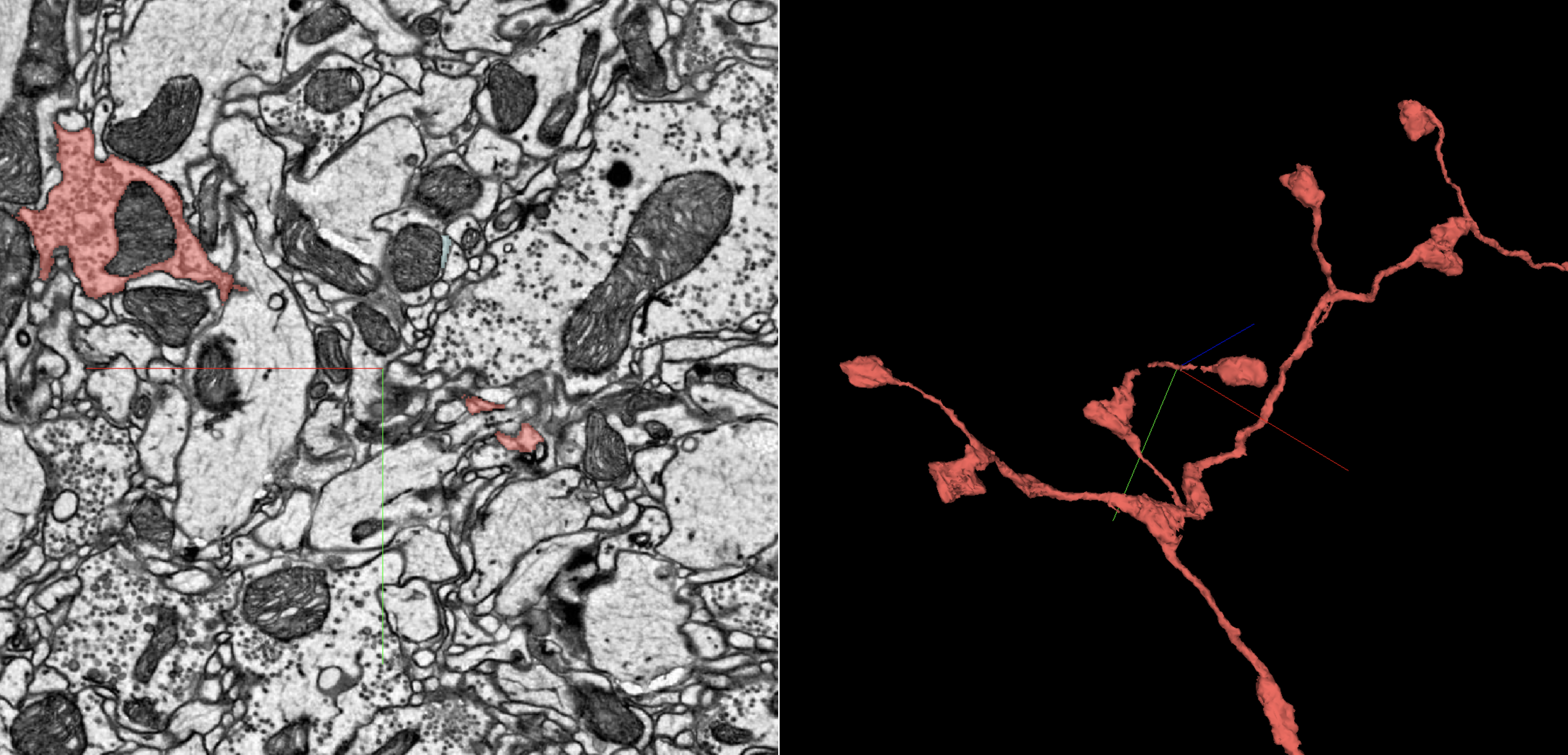
On the left hand side here you can see the way that a “floating” bouton may appear as a self-contained blob. To the right, note how this axonal terminal may seem rather small, but as with the dendritic example earlier, it’s complete.
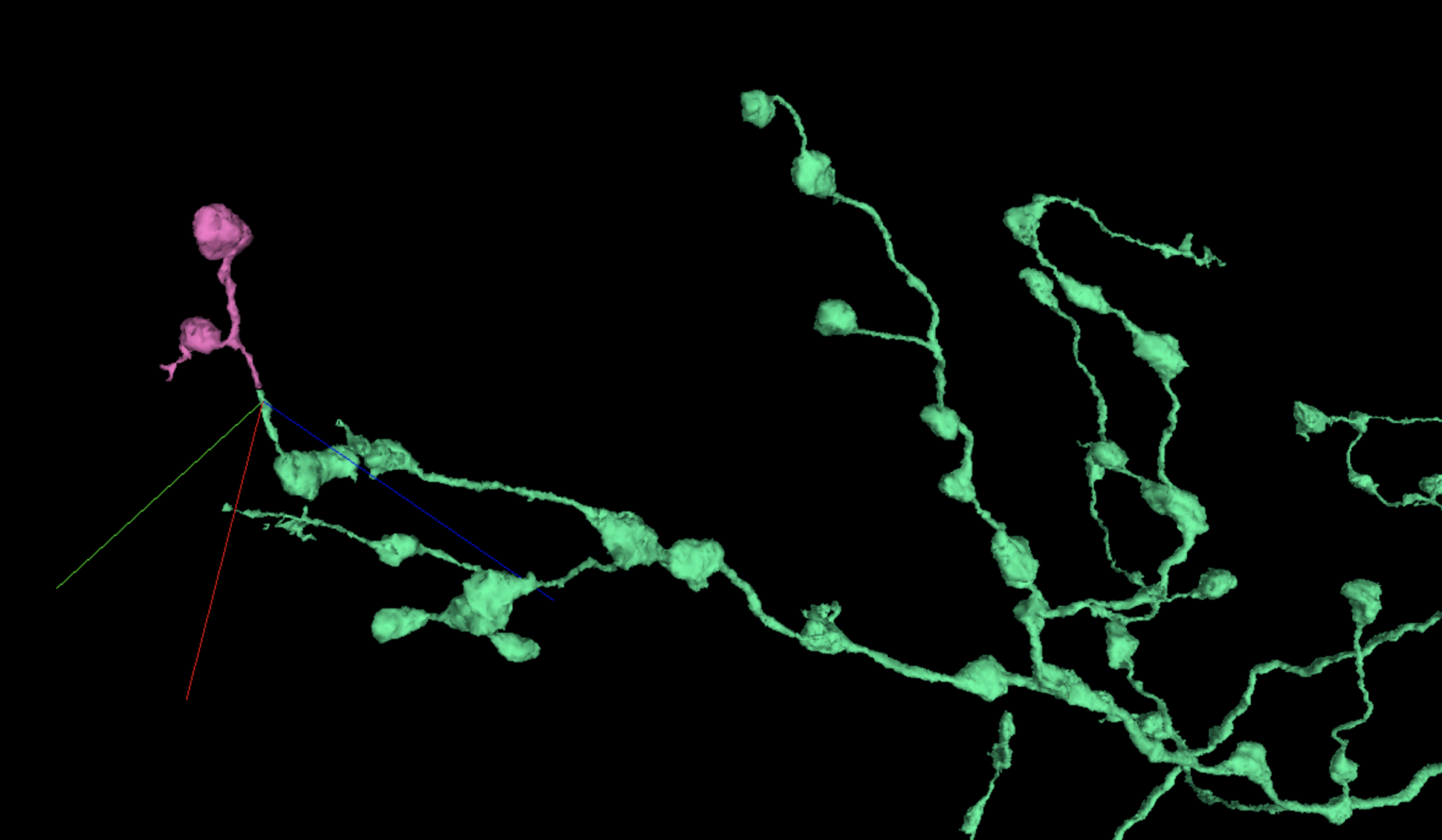
If you see an axonal branch with boutons at most of the end twigs, for consistency’s sake you should examine any twigs that seem to lack boutons, like in this example prior to merging:
Path Swaps
As with FlyWire, false continuations in the BANC dataset are common when branches are all aligned in the same direction and twisted closely around each other. The AI can also make this kind of mistake, known as a path swap, based on other errors like misalignments.
You can most easily identify a path swap by first looking at the 3D view and seeing a branch that extends similarly to an H-shaped, parallel merger — only the erroneous segment continues in one direction instead of two. Navigate there in the 2D to confirm the error; then you can merge the correct extension before splitting and removing the incorrect part.
This video for FlyWire can again be helpful for understanding path swaps in BANC. In fact, path swaps are especially common in BANC due to the parallel alignment of so many cells reaching between the central brain and the VNC.
Knowing Your Cell Is Complete
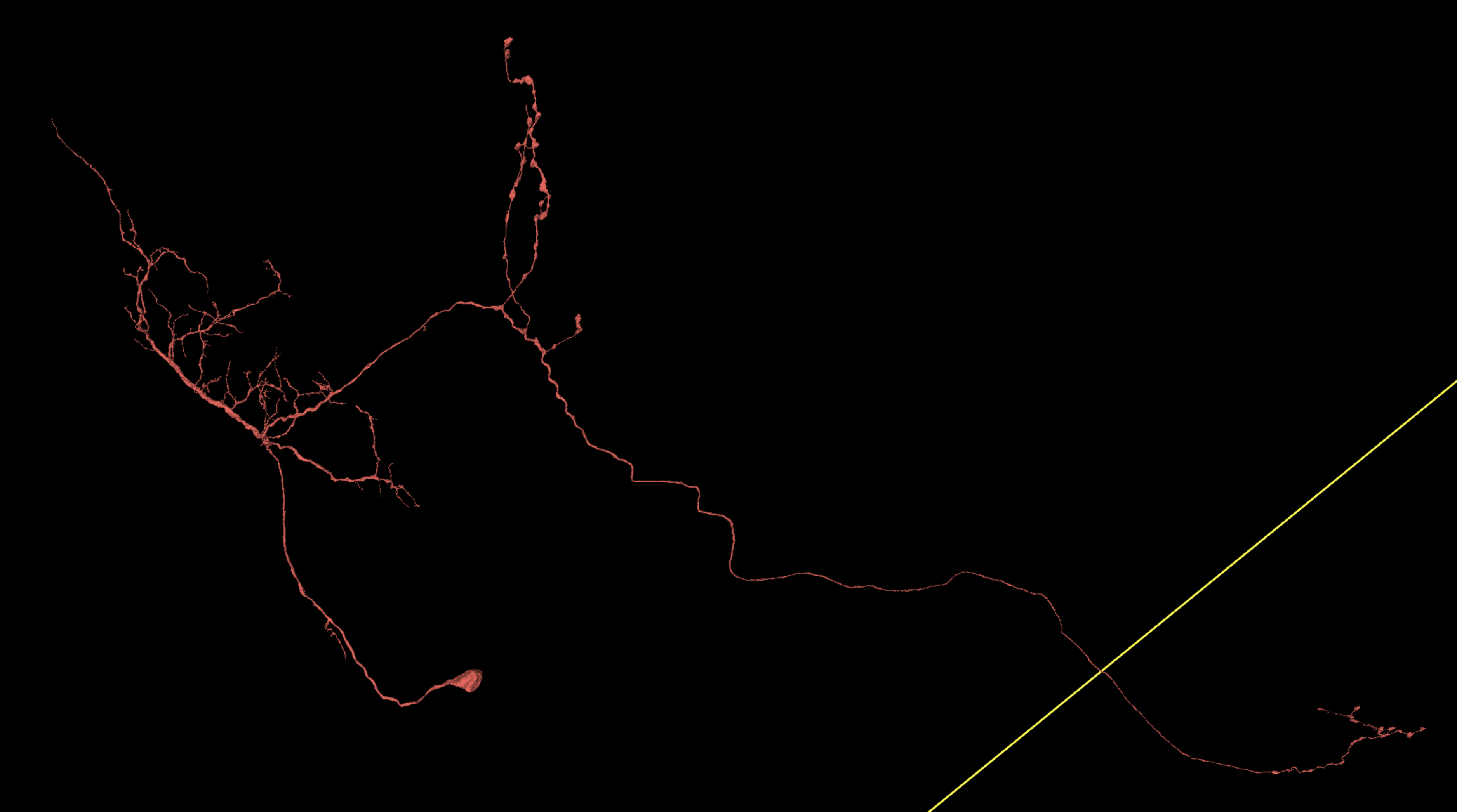
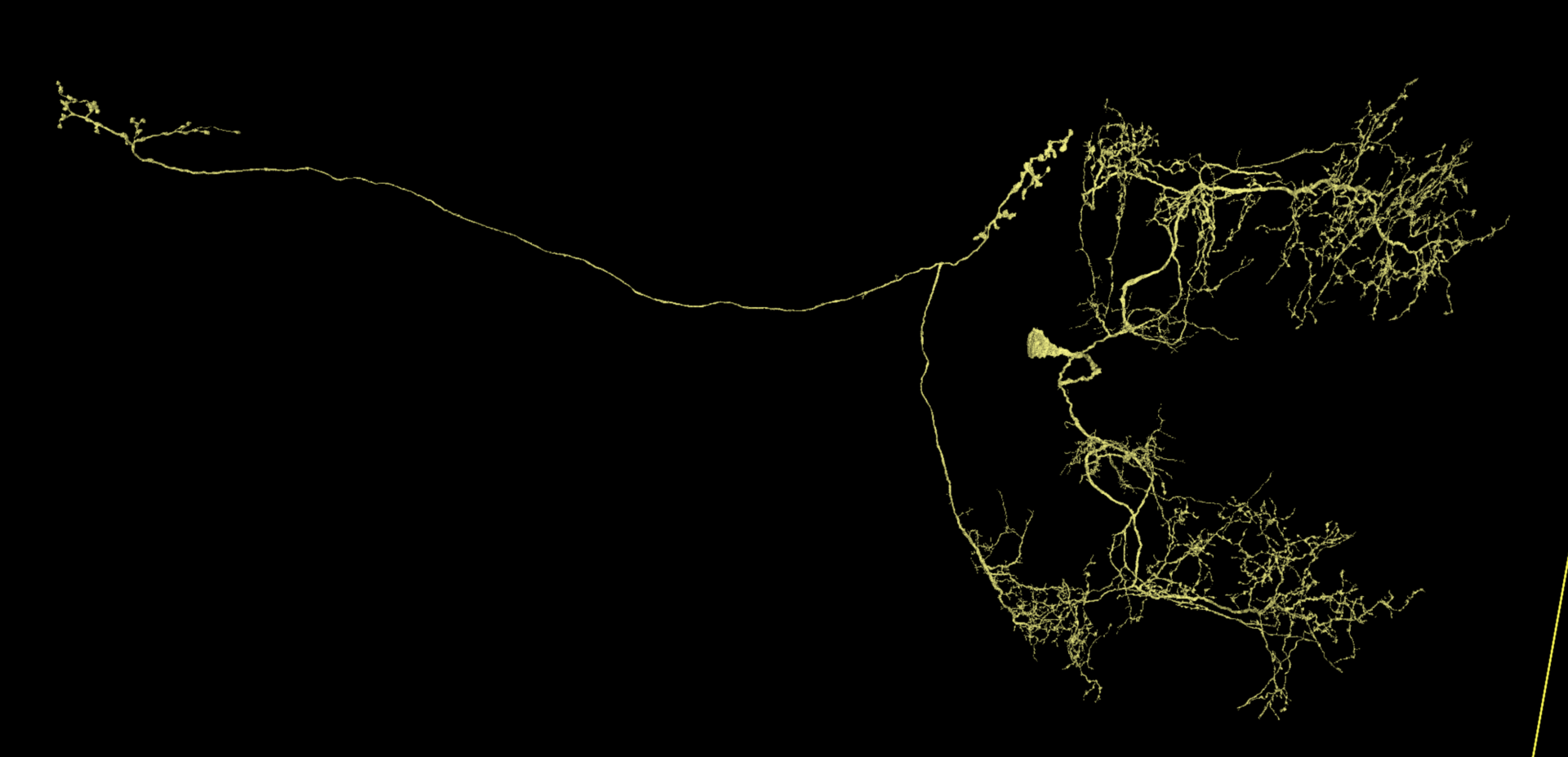
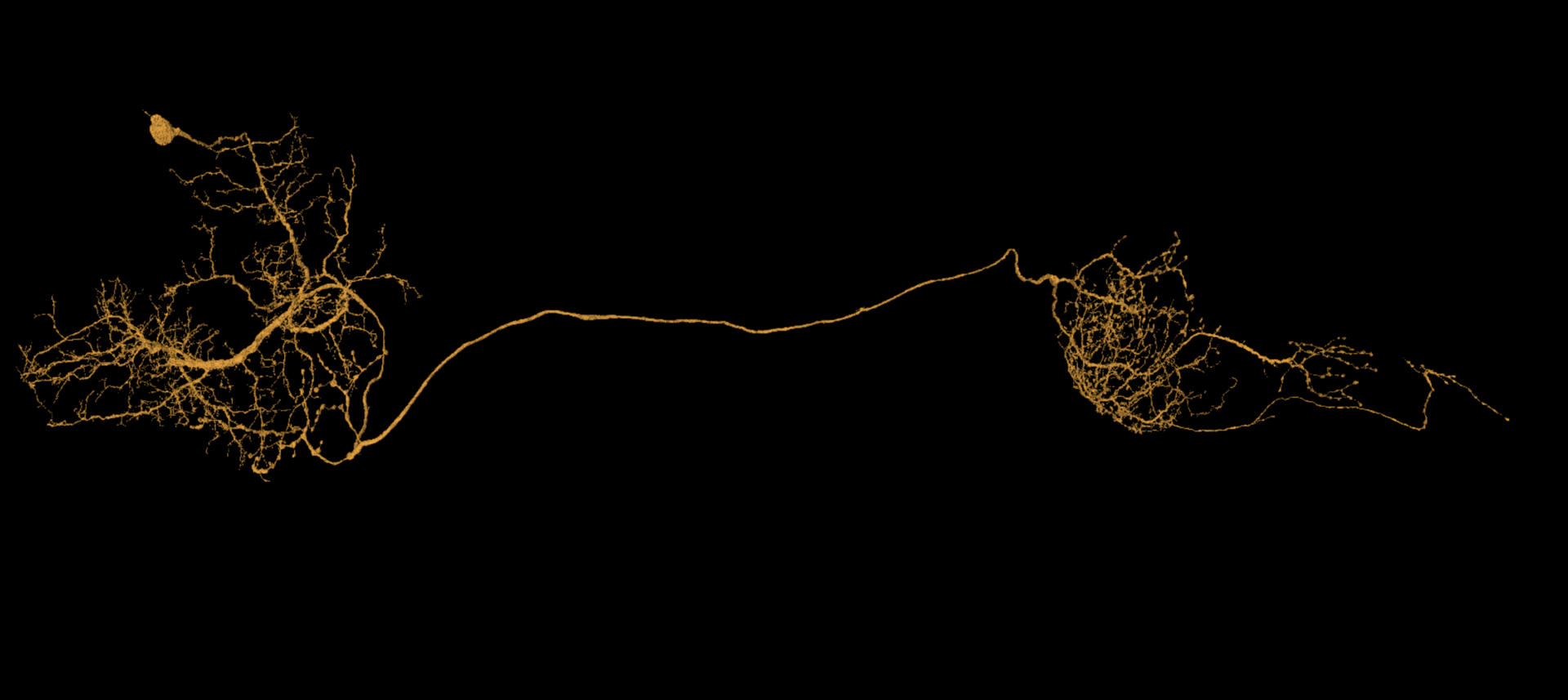
Your lab may have special expectations for when a cell should be considered adequately proofread. It depends on what data is being sought. However, if you’re trying to complete dendritic and axonal terminations in a reasonably thorough way, it can be challenging at first to recognize what the whole cell should look like when that process is finished.
Below are some examples of cells proofread within Seung Lab using a fairly thorough standard of completion. Notice how dendritic and axonal placement are variable by cell type. Also, as thorough as this is, not every single spine has been examined. If what you see already looks like this, then there probably isn’t much else that’s worth seeking out.
Found something else that’s tricky?
If you run into any questions or proofreading mysteries, feel free to send an email to flywire at princeton.edu or message #banc in FlyWire Slack.